01. Introduction
Heading
Introduction
ND320 C3 L2 01 Welcome To The Lesson & Overview
Summary of Overview + Lesson Outline
Conceptual map
Let’s look at our conceptual map again, highlighting the areas that we will cover in this lesson.
Conceptual Lesson Overview/Map
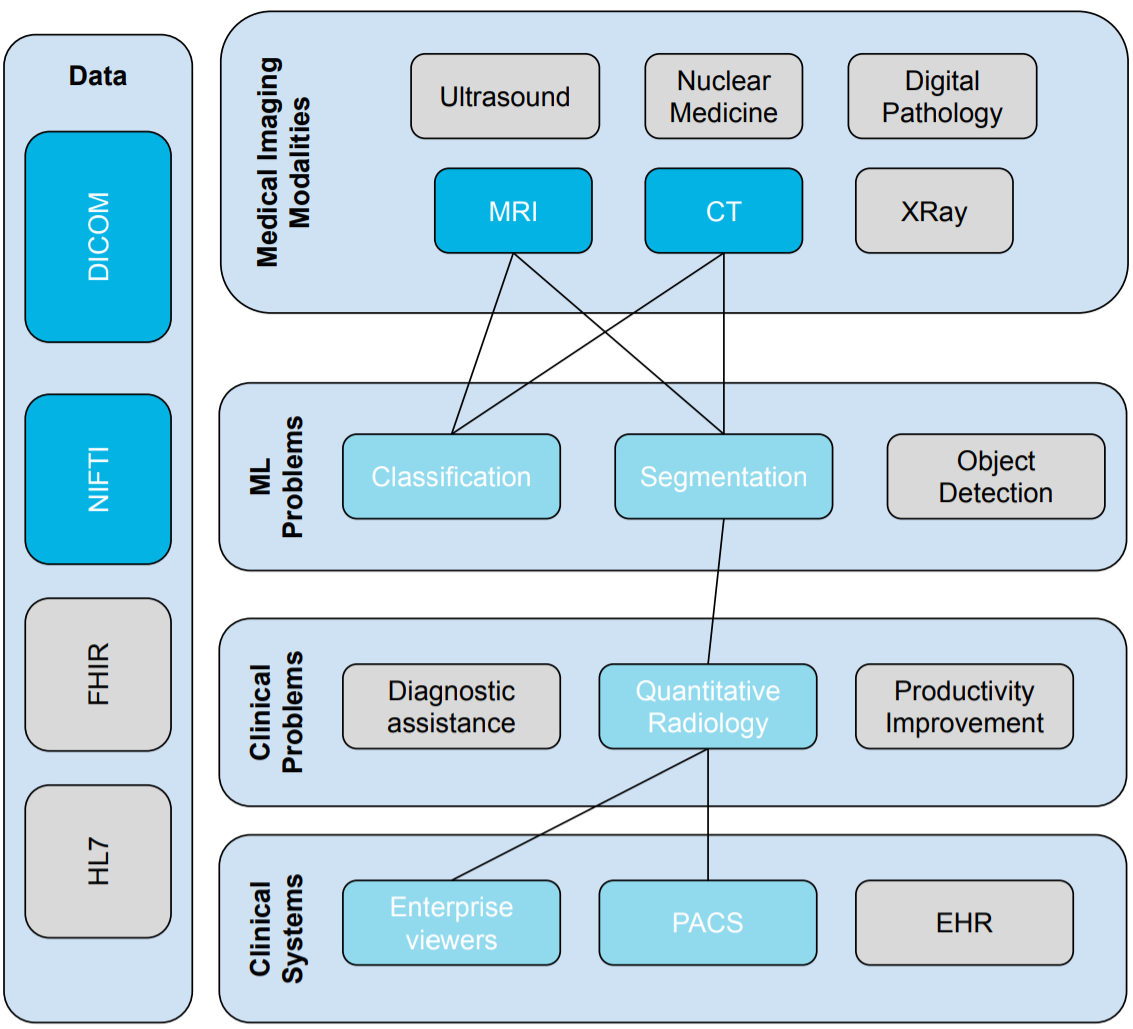
Lesson Overview (focus areas are highlighted with dark-blue color)
Lesson Summary
Lesson Summary
In this lesson we will cover the following:
- Basics of DICOM and NIFTI file formats
- A short introduction into viewing 3D medical images and some of the tools that you can use for viewing. This will be followed by an exercise where you will be asked to load a 2D DICOM image and convert it into PNG, applying proper windowing transform.
- Some important parameters to consider when analyzing datasets. This will be followed by an exercise where you will need to load a 3D volume and extract slices from it.
- A walkthrough of a Notebook with an approach to doing exploratory data analysis on a single 3D CT volume, and then an EDA for a dataset of multiple volumes. After that, you will be asked to do another exercise by working out an approach to an EDA of your own, curating a “dirty” DICOM volume dataset.
As you progress through the material, you may be wondering at times as to why you would need to know the intricacies of DICOM and NIFTI standards. I assure you that understanding how the standard is organized is crucial to understanding the data that you are dealing with. DICOM standard defines not only how pixels are stored, but also how the metadata is stored. A trove of valuable information is contained in DICOM metadata and this information could be used for all sorts of AI tasks - curating dataset, doing rapid QA, routing images to proper models, ensuring confidentiality compliance, etc. Also, if the content still seems too far removed, you might want to jump right into the exercises, and if they become confusing, come back to the lecture material, and use that to put you through.
So rock on!